New CRISPR tool targets RNA in mammalian cells
Researchers from the Broad Institute of MIT and Harvard have shown that a CRISPR-based editing system can cut and bind RNA in mammalian cells. In a paper out this week in Nature, the team used CRISPR-Cas13, which the researchers had helped discover, to both reduce RNA levels and "tag" RNAs in order to view and track them within cells. The researchers previously used CRISPR-Cas13 to target RNA in bacterial cells, but proving that the system could work safely and effectively in mammalian cells was a critical step toward using the system to study human biology and disease.
Having this sort of programmable tool for modulating RNA in mammalian cells creates new opportunities for learning how cells function and, potentially, for designing safer therapeutics. Unlike editing DNA, which makes permanent changes to the genome of a cell, targeting RNA could enable researchers to make temporary changes that alter the amount of protein produced by a gene rather than stopping production entirely.
"Even though we have good tools to delete genes, they still have many limitations that make the study of gene function difficult," explains co-first author Omar Abudayyeh, who is a graduate student in the lab of Broad core member and MIT associate professor Feng Zhang. "Cas13 allows you to bring down gene expression levels without completely eliminating them, which is useful for studying genes and may offer a less toxic therapeutic approach to correcting genetic diseases."
The team, led by scientists from Zhang's lab, tested Cas13 enzymes from fifteen different microbes to find the one, from Leptotrichia wadei (LwaCas13a), that was best-suited for the task. Using LwaCas13a enabled them to cut specific sites in targeted RNA with greater specificity than the current RNA-knockdown tool-of-choice, RNA-interference (RNAi). Although RNAi can be a useful tool, it often leads to unwanted off-target effects, making experiments difficult to interpret. Such off-target effects were significantly reduced with Cas13.
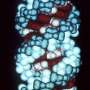
Zhang's team also demonstrated that a so-called "dead" variant of Cas13 that binds RNA but doesn't cut it can be combined with bright fluorescent "tags" to visually track target RNA as it moves within the cell.
"Our engineering of Cas13 here to bind and image transcripts shows the promise of this platform for the development of a broader set of tools to monitor and manipulate RNA," adds co-first author Jonathan Gootenberg, who is also a graduate student in Zhang's lab as well as the lab of Broad core member Aviv Regev.
The researchers note that CRISPR-Cas13's native ability to bind RNA also makes it easier to use than other technologies, which currently require researchers to modify the genome of an organism in order to create a binding site. These features could make CRISPR-Cas13 a key addition to biologists' toolbox for studying gene function; researchers can obtain CRISPR-Cas13 tools via the non-profit plasmid repository Addgene.
More information: Omar O. Abudayyeh et al. RNA targeting with CRISPR–Cas13, Nature (2017). DOI: 10.1038/nature24049
Journal information: Nature
Provided by Broad Institute of MIT and Harvard