October 17, 2017 report
Tiny protein coiled coils that self-assemble into cages
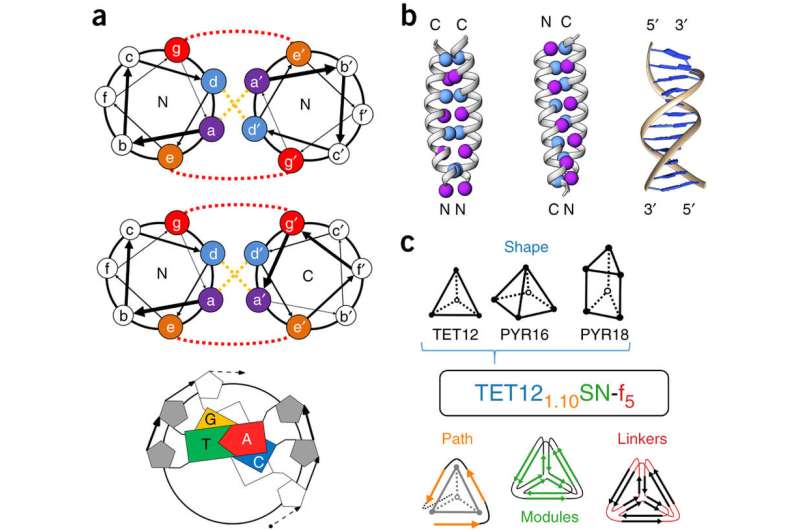
(Phys.org)—A large team of researchers with members from Slovenia, the U.K, Serbia, France and Spain has developed a technique that causes proteins to self-assemble into geometric shapes on demand. In their paper published in the journal Nature Biotechnology, the group describes their technique and possible uses for the tiny cages.
In recent years, scientists have manipulated DNA strands to cause them to bind together into useful shapes (DNA origami). In this new effort, the researchers have done something similar using proteins instead. Those in the field believe that such objects could be useful for applications like building packets for delivering drugs to targeted locations in the human body.
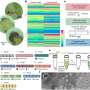
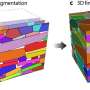
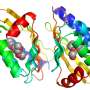
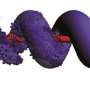
To cause the proteins to self-assemble, the researchers twisted dual strands of amino acid sections (coiled coils) into amino acid strands that were placed into cut sections of naturally occurring protein strands. The researchers describe the coiled coils as resembling yarn segments. The sections that were introduced into the chain were chosen specifically because prior testing had shown they would flex in a desired way when exposed to factors like electricity or water. The result was a single strand with bends that caused the overall strand to take the shape of a geometric object. The team reports that they were able to create tetrahedrons, four-sided pyramids and triangular prisms. Testing of the structures showed them to be soluble in aqueous solutions and that they could take their shapes both inside and outside of cells. The team also showed that the structures could be formed and possibly used in living mice.
The researchers note that parameters such as the charge of the coiled coils and the cap that forms at their ends can be adjusted to create different shapes and for dictating the conditions under which the proteins self-assemble. They further report that it was possible to create a tool-box of building blocks of the structures, allowing for larger, more complex structures from the basic shapes. They suggest such constructs could be used to carry drugs or vaccinations through the body or to create other structures that incorporate amino-acid functionality.
More information: Ajasja Ljubetič et al. Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo, Nature Biotechnology (2017). DOI: 10.1038/nbt.3994
Abstract
Polypeptides and polynucleotides are natural programmable biopolymers that can self-assemble into complex tertiary structures. We describe a system analogous to designed DNA nanostructures in which protein coiled-coil (CC) dimers serve as building blocks for modular de novo design of polyhedral protein cages that efficiently self-assemble in vitro and in vivo. We produced and characterized >20 single-chain protein cages in three shapes—tetrahedron, four-sided pyramid, and triangular prism—with the largest containing >700 amino-acid residues and measuring 11 nm in diameter. Their stability and folding kinetics were similar to those of natural proteins. Solution small-angle X-ray scattering (SAXS), electron microscopy (EM), and biophysical analysis confirmed agreement of the expressed structures with the designs. We also demonstrated self-assembly of a tetrahedral structure in bacteria, mammalian cells, and mice without evidence of inflammation. A semi-automated computational design platform and a toolbox of CC building modules are provided to enable the design of protein cages in any polyhedral shape.
Journal information: Nature Biotechnology
© 2017 Phys.org