Plant discovery opens frontiers

University of Adelaide researchers have discovered a biochemical mechanism fundamental to plant life that could have far-reaching implications for the multibillion dollar biomedical, pharmaceutical, chemical and biotechnology industries.
There are up to 80,000 fundamental-to-life enzymes working in plant or mammalian bodies, upon which almost all biochemical reactions depend. Enzymes carry out many chemical reactions, products of which can be used as building blocks or metabolites in a body, or they can serve as an energy source for every function in a body.
Researchers have discovered a new enzyme catalytic mechanism—catalysis being a process which speeds up chemical reactions—which they say could impact on work in biofuels production, in food and materials processing, and in drug discovery.
Their work has been published today in Nature Communications.
"The foundation for this was laid down in our earlier work but, at that time, we could not explain some fundamental processes at work in plants as these occur at immense rates," said lead researcher Professor Maria Hrmova from the University of Adelaide's School of Agriculture, Food and Wine.
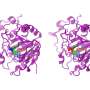
"The solution to this came up only recently, when we combined high-resolution X-ray crystallography, enzyme kinetics, mass spectrometry, nuclear magnetic resonance spectroscopy, and multi-scale 3-D molecular modelling tools to show those super-fast processes.
"Using computer simulations, we discovered a remarkable phenomenon during initial and final catalytic events near the surface of the plant glucose-processing enzyme. The enzyme formed a cavity which allowed the trapped glucose to escape to allow for the next round of catalysis," Professor Hrmova said.
"Now that we can simulate these nanoscale movements we have opened the door to new knowledge on enzyme dynamics that was inaccessible before. These discoveries are made once in a generation."
Professor Hrmova said the discovery of the catalytic mechanism involved specialists in plant and molecular biology, biochemistry, biophysics, bioinformatics and computer science from seven countries (Australia, France, Thailand, Spain, Chile, Slovak Republic and China).
"Being able to describe these minute and ultra-fast processes—unable to be captured via usual experimental techniques—means we can now work to improve enzyme catalytic rates, stability and product inhibition. This will have significance in biotechnologies to develop or manufacture products through novel forms of bioengineered enzymes that could be also used outside of biological systems."
More information: Victor A. Streltsov et al. Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site, Nature Communications (2019). DOI: 10.1038/s41467-019-09691-z
Journal information: Nature Communications
Provided by University of Adelaide